CONTACT
-
Department of Biological Sciences
Email
718-960-8235
Davis Hall, Room 217Office Hours
Monday - Friday, 9:00am-5:00pm
Faculty Haiping Cheng
 E-mail address: haiping.cheng@lehman.cuny.edu
E-mail address: haiping.cheng@lehman.cuny.edu
Phone Number: 718-960-7190
Office: Science Hall 3408 (Lab: Science Hall 3411, 718-960-7227)
Rank: Professor
Degrees and Sources of Degrees: B.A, M.A., Fudan Univ., China; Ph.D., Univ. of Mass., Amherst; Postdoc., Mass. Institute of Technology
Research interests: Antibiotics discovery: A few years ago, we accidentally identified a set of unique conditions that can trigger commensal bacteria into producing bacterial culture supernatants with broad-spectrum antibiotic activities, including against the difficult to kill-Mycobacterium. We are currently focusing on identifying the cellular targets of this antibiotic activity using molecular biology approaches and identifying the antibiotic compound(s) in the supernatant using biochemical approaches.
Antifungals discovery: Yeast is part of a group of fungi which includes yeast, mushrooms, and mold that share our living environment. The fact that fungi share basic structures with human cells makes it difficult to find antifungals that can kill or inhibit fungi growth without harming human cells. This is the reason why there are limited number of antifungal drugs on the market. We have started a project to search for compounds that can inhibit the growth of yeast by triggering programmed cell death.
Bacterial host sensing mechanism: Bacteria have to sense the presence of their natural hosts and turn on the production of special factors that will enable the invasion of their natural hosts. Using the nitrogen-fixing symbiotic relationship between the Gram-negative soil bacterium, Sinorhizobium meliloti, and its plant host, alfalfa, as a model, we are studying the signaling function of a molecular switch, the RSI host invading switch and search for the signals sensed the RSI switch.
The global spread of antibiotic-resistant bacteria has led the WHO and CDC to declare a worldwide emergency and call for an effort to develop new antibiotics against multi-drug-resistant pathogens. The WHO reports a lack of significant development of novel antibiotics that are effective against multi-drug resistant strains. Antibiotic-resistant bacterial infections are responsible for 64.5 million yearly infections with an annual mortality rate of 812,000 worldwide, and 35,000 deaths occurring in the US. While finding a novel antibiotic has been a global effort for decades, it remains a challenge due to the lack of new cellular targets and new compounds with antibiotic activity. Good cellular targets for antibiotics are bacterial proteins (protein complex) or biological processes that are present only in bacteria and absent in human cells. Despite joint efforts from scientists worldwide, there has been little progress in antibiotic discovery in the last 50 years, which is a true reflection of the difficulties in finding new antibiotics and drug targets. In our search for new antibiotics, we have developed a reproducible method to produce a bacterial-derived culture supernatant with strong and broad-spectrum antibiotic activity (active supernatant) or without antibiotic activity (inactive supernatant). Unlike bacteriocins and microcins, our active supernatant inhibits the growth of the producer strain and all the bacteria that we have tested thus far, including clinical isolates that we obtained from the NIH funded Antibiotic Resistance Leadership Group (ARLG). Preliminary results suggest that our active supernatant is not toxic to human, rat, and yeast cells. We are currently focusing on identifying the cellular targets of this antibiotic activity using molecular biology approaches and identifying the antibiotic compound(s) in the supernatant using biochemical approaches.
Recent publications
Ella Vardeman, Hai-Ping Cheng, Ina Vandebroek, and Edward Kennelly. 2024 Caribbean medicinal plant Argemone mexicana L.: metabolomic analysis and in vitro effect on vaginal microbiota. Journal of Ethnopharmacology. 337(2025)118830. PMID:39277064
O’Connor, N.A.; Syed, A.; Kastrat, E.; Cheng, H.-P. 2024. Antibacterial Silver Nanoparticle Containing Polydopamine Hydrogels That Enhance Re-Epithelization. Gels 10, 363. https://doi.org/10.3390/
Ertan Kastrat and Hai-Ping Cheng. 2024. Escherichia coli has an undiscovered ability to inhibit the growth of both Gram-negative and Gram-positive bacteria. Scientific Report 14, 7420. PMID: 38548840. https://doi.org/10.1038/s41598-024-57996-x
Esteban Chaves-Olarte, Jazmín Meza-Torres, Fabiola Herrera-Rodríguez, Esteban Lizano-González, Marcela Suárez-Esquivel, Kate S. Baker, Olga Rivas-Solano, Nazareth Ruiz-Villalobos, Fabián Villalta-Romero, Hai-Ping Cheng, Graham C. Walker, Axel Cloeckaert, Nicholas R. Thomson, Teresa Frisan, Edgardo Moreno, Caterina Guzmán-Verri. 2023 A sensor histidine kinase from a plant-endosymbiont bacterium restores the virulence of a mammalian intracellular pathogen. Microbial Pathogenesis. 185:106442. PMID: 37944675.
Binsheng Luo, Ertan Kastrat, Taylan Morcol, Haiping Cheng, Edward Kennelly, and Chunlin Long. 2021. Gaultheria longibracteolata, an alternative source of wintergreen oil. Food Chemistry. 16:342 (Epub 2020 Oct. 9.) PMID:33097325
Fungal infections account for approximately 1.5 million annual deaths globally. Scientists are searching for unique targets for drug development to overcome the growing prevalence of resistance. The emergence of resistance for the three classes of antifungals; azoles, echinocandins, and polyenes, severely limits treatment options. Given the poor ability of antifungal drugs to distinguish between fungal pathogens and host cells, the emergence of antifungal resistance, and the recently discovered multidrug-resistant (MDR) Candia auris, it is reasonable to predict that fungi are entering the post-antibiotic era seen with bacteria. There is an urgent need for new targets for therapeutics, especially those with fundamental physiological importance, to prevent the rapid development of resistance. Additionally, the recently discovered association between fungi and cancer, and the known similarities in yeast and cancer cell metabolism, justify the importance of expanding on the understanding of programmed cell death in yeast. Like bacteria, yeast also produced a variety of metabolites to control their population dynamics. Our studies on yeast programmed cell death have led us to reproducibly collect supernatants from yeast cultures with strong fungicidal activity against a range of yeasts, including Candida albicans and Candida auris by triggering, what we suspect to be, programmed cell death. We are currently employing biochemical approaches to identify and isolate the active compound(s) in the supernatant and molecular approaches to understand the mechanism by which this inhibitory compound(s) is produced.
Recent publications
Parbhudayal, Raveena, and Hai-Ping Cheng. "Exploring sugar-induced cell death in yeast: implications for diabetes and cancer research." Frontiers in Cell Death 4 (2025): 1470093.
Sinorhizobium meliloti is a Gram-negative bacterium that can live freely in the soil or establish a nitrogen-fixing symbiosis with its plant host alfalfa (Medicago sativa) through a complex invasion process, which requires it to change from a free-living form to host-invading form. Once the invading S. meliloti cells reach alfalfa root nodules and they differentiate into their nitrogen-fixing form, bacteroid. Different genes will be expressed by S. meliloti for different forms of living to maximize its growth potential. This change is best represented by the free-living S. meliloti production of flagella for motility in its natural soil environment and by the host-invading S. meliloti production of succinoglycan, a bacterial exopolysaccharide, required for the invasion of host plant alfalfa for the establishment of nitrogen-fixing symbiosis.
The switch of S. meliloti cells from free-living to host-invading is controlled by a 3-protein complex consisting of the periplasmic ExoR protein, membrane sensor ExoS protein, and transcriptional regulator ChvI protein. We named this protein complex the RSI Host Invading Switch, the RSI Switch for short. The ExoR, ExoS, and ChvI proteins have been found in more than 100 different bacteria including animal and plant pathogens, and plant symbionts, suggesting that the RSI Switch might be part of a common molecular switch controlling the transformation of bacterial cells from free-living form to host-invading form.
As shown in the diagram of our current model RSI Switch, the ExoR protein plays an essential role in determining the ON/OFF status of the RSI Switch. The ExoR is first synthesized in the precursor form, ExoRp, exported to periplasm without its signal peptide to form the mature and active form, ExoRm., which can either form a complex with ExoS sensor or subject to digestion by a yet to identified periplasmic protease to form non-functional ExoRc20, So the levels periplasmic ExoRm is kept in equilibrium by the combination of synthesis and digestion. When ExoRm is maintained at medium levels, the RSI Switch is in the OFF state, at which there are enough ExoRm to form a complex with ExoS and keep ExoS in the inactive state so that ChvI is in the non-phosphorylated and inactive state. The reduction of ExoRm levels turns ON the RSI Switch, during which the ExoS sensor becomes free of ExoRm and active, phosphorylates ChvI, and then activates the expression of succinoglycan synthesis genes and suppresses the flagella genes.
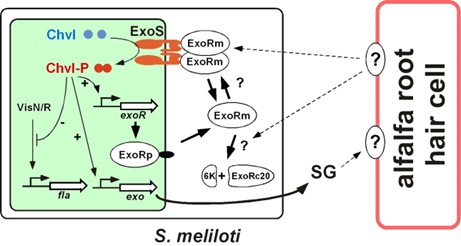
We are currently working on identifying the host signals, characterizing the ExoRm -ExoS interaction, and the ExoRm proteolysis, to improve our understanding of the molecular mechanism of the Sinorhizobium meliloti host invasion switch, which controls the change of free-living S. meliloti cells to host-invading S. meliloti cells.
- Esteban Chaves-Olarte, Jazmín Meza-Torres, Fabiola Herrera-Rodríguez, Esteban Lizano-González, Marcela Suárez-Esquivel, Kate S. Baker, Olga Rivas-Solano, Nazareth Ruiz-Villalobos, Fabián Villalta-Romero, Hai-Ping Cheng, Graham C. Walker, Axel Cloeckaert, Nicholas R. Thomson, Teresa Frisan, Edgardo Moreno, Caterina Guzmán-Verri. 2023 A sensor histidine kinase from a plant-endosymbiont bacterium restores the virulence of a mammalian intracellular pathogen. Microbial Pathogenesis. 185:106442. [Link]
- Eliza M. Wiech, Hai-Ping Cheng, and Shaneen M. Singh. 2015 Computational analyses of the Sinorhizobium meliloti Rm1021 ExoR protein. Protein Chem. 24(3):319-327. [PDF]
- Mary Ellen Heavner, Wei-Gang Qiu, and Hai-Ping Cheng. 2015 Phylogenetic Co-Occurrence of ExoR, ExoS, and ChvI, Components of the RSI Bacterial Invasion Switch, Suggests a Key Adaptive Mechanism Regulating the Transition between Free-Living and Host-Invading Phases in Rhizobiales. PLoS One.2015 26;10(8):e0135655. [PDF]
- Myrthala M Verástegui-Valdés; Yu-Jing Zhang; Flor N Rivera-Orduña; Hai-Ping Cheng; Xin-Hua Sui; En-Tao Wang. 2014. Microsymbionts of Phaseolus vulgaris in acid and alkaline soils of Mexico. Systematic and Applied Microbiology. 37: 605-612. [PDF]
- Hai-Yang Lu, Li Luo, Meng-Hua Yang, and Hai-Ping Cheng. 2012. Sinorhizobium meliloti ExoR is the target of periplasmic proteolysis. J. Bacteriology. 194(15): 4029-4040 [PDF]
- Hai-Yang Lu and Hai-Ping Cheng. 2010. Autoregulation of Sinorhizobium meliloti exoR gene expression. Microbiology 156: 2092-2101 [PDF]
- Shi-Hua Shen, Feng Chi, Kuixian Ji, Yu-xiang Jing, Ming-Feng Yang, Hai-Ping Cheng, Frank B. Dazzo. 2010. New ecological dynamics of GFP-tagged rhizobia after inoculation of tobacco plants. Journal of Microbiology and Biotechnology, 20(2): 238-244 [PDF]
- Xue-Song Zhang and Hai-Ping Cheng. 2006. Identification of Sinorhizobium meliloti early symbiotic genes by use of a positive functional screen. Applied Environmental Microbiology. 72:2738-2748 [PDF]
- Feng Chi, Shi-Hua Shen, Hai-Ping Cheng, Yu-Xiang Jing and Frank B. Dazzo. 2005. Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology. Applied Environmental Microbiology, 71:7271-7278 [PDF]
- Li Luo, Shi-Yi Yao, Anke Becker, Silvia Ruberg, Guan-Qiao Yu, Jia-Bi Zhu, and Hai-Ping Cheng. 2005. Identification of two new Sinorhizobium meliloti lysR like transcriptional regulators required for nodulation. J. Bacteriology, 187: 4562-4572 [PDF]
- Li Luo, Ming-Sheng Qi, Shi-Yi Yao, Hai-Ping Cheng, Jia-Bi Zhu, Shan-Jiong Shen, and Guan-Qiao Yu. 2005. Characterization of OxyR homolog from Sinorhizobium meliloti Rm1021 regulating the expression of catalase genes. Acta Biochimica et Biophysica Sinica, 37: 421-428 [PDF]
- Shi-Yi Yao, Li Luo, Katherine Har, Anke Becker, Silvia Ruberg, Guan-Qiao Yu, Jia-Bi Zhu, and Hai-Ping Cheng. 2004. The Sinorhizobium meliloti ExoR and ExoS proteins regulate both succinoglycan and flagella production. J. Bacteriology, 186:6042-6049 [PDF]
- Hai-Ping Cheng and Shi-Yi Yao. 2004. The promoters of the Sinorhizobium meliloti exoY gene and their expression during nodulation. FEMS. Microbiology Letters, 231:131-136 [PDF]
- Brett Pellock, Hai-Ping Cheng, and Graham C. Walker. 2000. Alfalfa root nodule invasion efficiency is dependent on Sinorhizobium meliloti. J. Bacteriology, 182:4310-4318 [PDF]
- Hai-Ping Cheng and Graham C. Walker. 1998. Succinoglycan production of Rhizobium meliloti 1021 is regulated by the ExoS/ChvI two-component regulatory system. J. Bacteriology. 180: 20-26 [PDF]
- Hai-Ping Cheng and Graham C. Walker. 1998. Succinoglycan is required for the formation of infection threads during the nodulation of alfalfa by Rhizobium meliloti 1021. J. Bacteriology. 180: 5183-5191 [PDF]
- Anke Becker, S. Ruber, H. Kuster, A.A. Roxlau, M. Keller, T. Ivashina, Hai-Ping Cheng, G.C. Walker, and A. Pulher. 1997. The 32-Kilobase exp gene cluster of Rhizobium meliloti directing the biosynthesis of galactoglucan: genetic organization and properties of the encoded gene products. J. Bacteriology. 179:1375-1384. [PDF]
- Hai-Ping Cheng and Thomas G. Lessie. 1994. Multiple replicons constituting the genome of Pseudomonas cepacia 17616. J. Bacteriology.176:4034-4342 [PDF]
-
Department of Biological Sciences
Email
718-960-8235
Davis Hall, Room 217Office Hours
Monday - Friday, 9:00am-5:00pm
- See all contacts